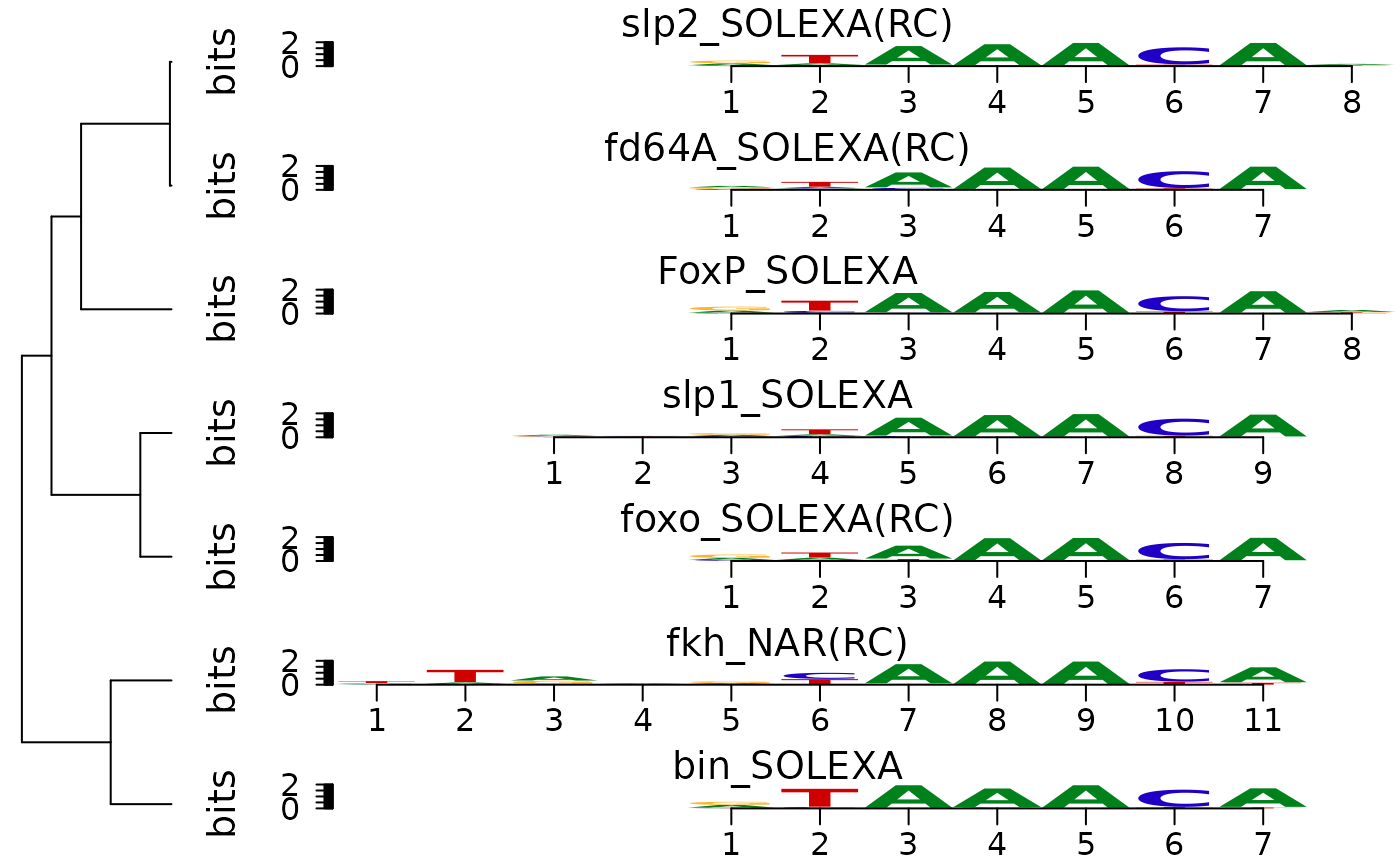
plot sequence logos stack with hierarchical cluster tree
plotMotifLogoStackWithTree.Rdplot sequence logos stack with hierarchical cluster tree
plotMotifLogoStackWithTree(pfms, hc, treewidth = 1/8, trueDist = FALSE, ...)
Arguments
| pfms | a list of position frequency matrices, pfms must be a list of class pfm |
|---|---|
| hc | an object of the type produced by stats::hclust |
| treewidth | the width to show tree |
| trueDist | logical flags to use hclust height or not. |
| ... | other parameters can be passed to plotMotifLogo function |
Value
none
Examples
#####Input##### pcms<-readPCM(file.path(find.package("motifStack"), "extdata"),"pcm$") #####Clustering##### hc <- clusterMotifs(pcms) ##reorder the motifs for plotMotifLogoStack motifs<-pcms[hc$order] motifs <- lapply(motifs, pcm2pfm) ##do alignment motifs<-DNAmotifAlignment(motifs) ##plot stacks plotMotifLogoStack(motifs, ncex=1.0)#> NULLplotMotifLogoStackWithTree(motifs, hc=hc)#> NULL