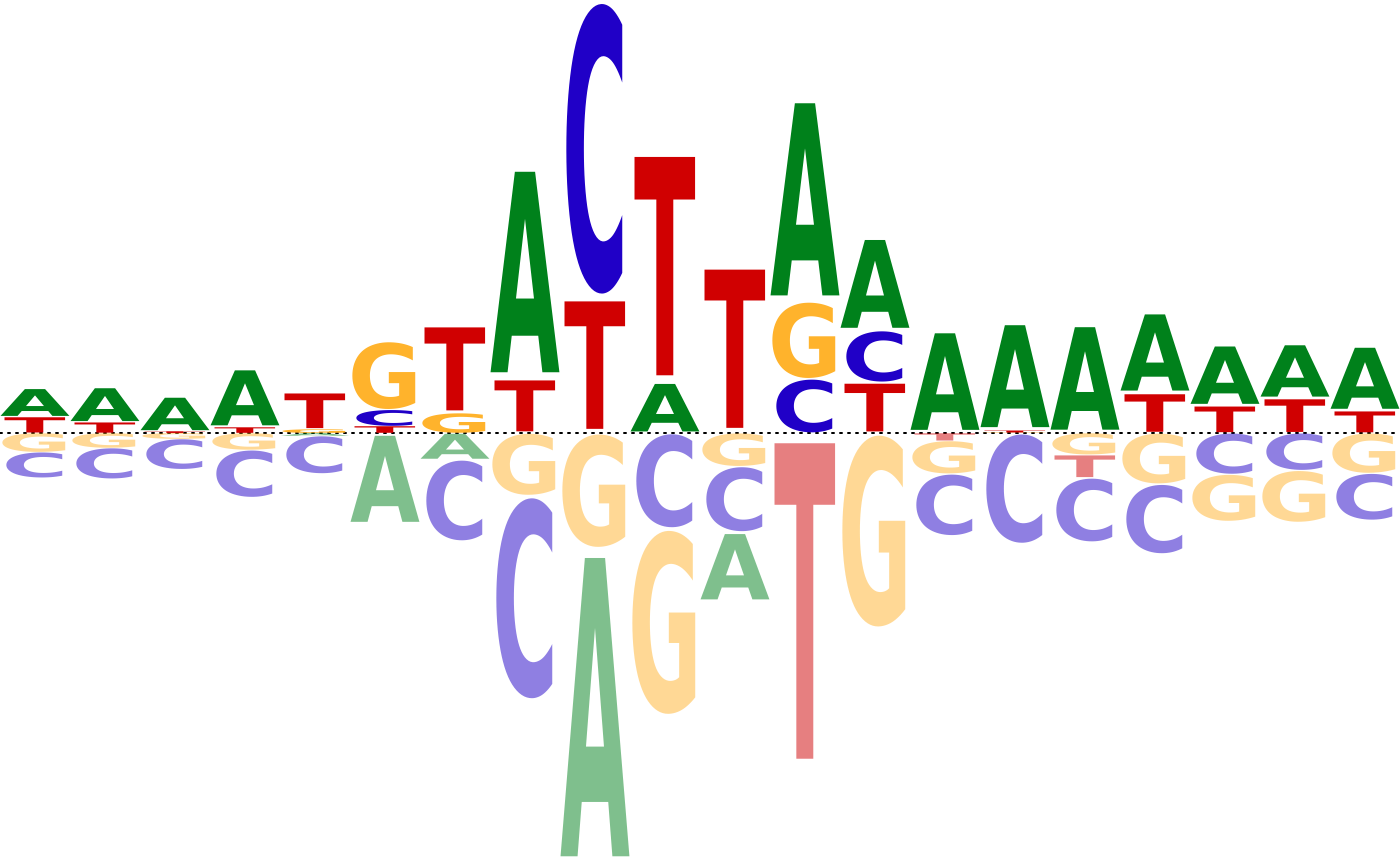Class "psam"
psam-class.RdAn object of class "psam" represents the position specific affinity
matrix (PSAM) of a DNA/RNA/amino-acid sequence motif. The entry stores a
matrix, which in row i, column j gives the affinity of observing
nucleotide/or amino acid i in position j of the motif.
methods for psam objects.
# S4 method for psam $(x, name) # S4 method for psam,ANY plot(x, y = "missing", ...) # S4 method for psam matrixReverseComplement(x) # S4 method for psam,numeric,logical addBlank(x, n, b) # S4 method for psam as.data.frame(x, row.names = NULL, optional = FALSE, ...) # S4 method for psam format(x, ...)
Arguments
| x | An object of class |
|---|---|
| name | Slot name. |
| y | Not use. |
| ... | Further potential arguments passed to |
| n | how many spaces should be added. |
| b | logical value to indicate where the space should be added. |
| row.names, optional | see as.data.frame |
Objects from the Class
Objects can be created by calls of the form
new("psam", mat, name, alphabet, color).
Methods
- addBlank
signature(x="psam", n="numeric", b="logical")add space into the position specific affinity matrix for alignment. b is a bool value, if TRUE, add space to the 3' end, else add space to the 5' end. n indicates how many spaces should be added.- matrixReverseComplement
signature(x = "psam")get the reverse complement of position specific affinity matrix.- plot
signature(x = "psam")Plots the affinity logo of the position specific affinity matrix.- $, $<-
Get or set the slot of
psam-class- as.data.frame
convert
psam-classto a data.frame- format
return the name_pfm of
psam-class
Examples
motif <- importMatrix(file.path(find.package("motifStack"), "extdata", "PSAM.mxr"), format="psam")[[1]] plot(motif)motif <- importMatrix(file.path(find.package("motifStack"), "extdata", "PSAM.mxr"), format="psam")[[1]] matrixReverseComplement(motif)#> An object of class "psam" #> Slot "mat": #> [,1] [,2] [,3] [,4] [,5] [,6] [,7] #> A 0.8876650 0.9460106 0.9100964 0.8915726 0.6913035 0.7429453 0.7344307 #> C 0.7454561 0.7424521 0.7393219 0.6914964 0.6939664 0.7396342 0.6842762 #> G 0.7317264 0.7736245 0.7516285 0.6555763 0.6158464 0.5410013 0.6308472 #> T 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 #> [,8] [,9] [,10] [,11] [,12] [,13] [,14] #> A 0.8877684 0.2172802 1.0000000 1.0000000 0.6368903 0.6408147 1.0000000 #> C 0.4445597 0.7007176 0.5686857 0.3067372 0.3136375 0.4615689 0.8262161 #> G 0.8895028 0.6566760 0.5194728 0.3987222 1.0000000 0.3081641 0.6216643 #> T 1.0000000 1.0000000 0.5136795 0.6013992 0.1751474 1.0000000 0.7255969 #> [,15] [,16] [,17] [,18] [,19] [,20] #> A 0.8424167 1.0000000 0.8634302 0.9127193 0.9350338 0.9662157 #> C 1.0000000 0.9118593 0.8102359 0.8935170 0.8701648 0.8755225 #> G 0.8642716 0.8119189 0.7437658 0.8342023 0.8321566 0.8596980 #> T 0.6393438 0.8952272 1.0000000 1.0000000 1.0000000 1.0000000 #> #> Slot "name": #> [1] "PSAM.mxr" #> #> Slot "alphabet": #> [1] "DNA" #> #> Slot "color": #> A C G T #> "#00811B" "#2000C7" "#FFB32C" "#D00001" #> #> Slot "tags": #> list() #> #> Slot "markers": #> list() #>#> An object of class "psam" #> Slot "mat": #> [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] #> A 0 1.0000000 1.0000000 1.0000000 1.0000000 0.8952272 0.6393438 0.7255969 #> C 0 0.8596980 0.8321566 0.8342023 0.7437658 0.8119189 0.8642716 0.6216643 #> G 0 0.8755225 0.8701648 0.8935170 0.8102359 0.9118593 1.0000000 0.8262161 #> T 0 0.9662157 0.9350338 0.9127193 0.8634302 1.0000000 0.8424167 1.0000000 #> [,9] [,10] [,11] [,12] [,13] [,14] [,15] #> A 1.0000000 0.1751474 0.6013992 0.5136795 1.0000000 1.0000000 1.0000000 #> C 0.3081641 1.0000000 0.3987222 0.5194728 0.6566760 0.8895028 0.6308472 #> G 0.4615689 0.3136375 0.3067372 0.5686857 0.7007176 0.4445597 0.6842762 #> T 0.6408147 0.6368903 1.0000000 1.0000000 0.2172802 0.8877684 0.7344307 #> [,16] [,17] [,18] [,19] [,20] [,21] #> A 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 #> C 0.5410013 0.6158464 0.6555763 0.7516285 0.7736245 0.7317264 #> G 0.7396342 0.6939664 0.6914964 0.7393219 0.7424521 0.7454561 #> T 0.7429453 0.6913035 0.8915726 0.9100964 0.9460106 0.8876650 #> #> Slot "name": #> [1] "PSAM.mxr" #> #> Slot "alphabet": #> [1] "DNA" #> #> Slot "color": #> A C G T #> "#00811B" "#2000C7" "#FFB32C" "#D00001" #> #> Slot "tags": #> list() #> #> Slot "markers": #> list() #>#> An object of class "psam" #> Slot "mat": #> [,1] [,2] [,3] [,4] [,5] [,6] [,7] #> A 1.0000000 1.0000000 1.0000000 1.0000000 0.8952272 0.6393438 0.7255969 #> C 0.8596980 0.8321566 0.8342023 0.7437658 0.8119189 0.8642716 0.6216643 #> G 0.8755225 0.8701648 0.8935170 0.8102359 0.9118593 1.0000000 0.8262161 #> T 0.9662157 0.9350338 0.9127193 0.8634302 1.0000000 0.8424167 1.0000000 #> [,8] [,9] [,10] [,11] [,12] [,13] [,14] #> A 1.0000000 0.1751474 0.6013992 0.5136795 1.0000000 1.0000000 1.0000000 #> C 0.3081641 1.0000000 0.3987222 0.5194728 0.6566760 0.8895028 0.6308472 #> G 0.4615689 0.3136375 0.3067372 0.5686857 0.7007176 0.4445597 0.6842762 #> T 0.6408147 0.6368903 1.0000000 1.0000000 0.2172802 0.8877684 0.7344307 #> [,15] [,16] [,17] [,18] [,19] [,20] [,21] [,22] [,23] #> A 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 0 0 0 #> C 0.5410013 0.6158464 0.6555763 0.7516285 0.7736245 0.7317264 0 0 0 #> G 0.7396342 0.6939664 0.6914964 0.7393219 0.7424521 0.7454561 0 0 0 #> T 0.7429453 0.6913035 0.8915726 0.9100964 0.9460106 0.8876650 0 0 0 #> #> Slot "name": #> [1] "PSAM.mxr" #> #> Slot "alphabet": #> [1] "DNA" #> #> Slot "color": #> A C G T #> "#00811B" "#2000C7" "#FFB32C" "#D00001" #> #> Slot "tags": #> list() #> #> Slot "markers": #> list() #>#> [,1] [,2] [,3] [,4] [,5] [,6] [,7] #> A 1.0000000 1.0000000 1.0000000 1.0000000 0.8952272 0.6393438 0.7255969 #> C 0.8596980 0.8321566 0.8342023 0.7437658 0.8119189 0.8642716 0.6216643 #> G 0.8755225 0.8701648 0.8935170 0.8102359 0.9118593 1.0000000 0.8262161 #> T 0.9662157 0.9350338 0.9127193 0.8634302 1.0000000 0.8424167 1.0000000 #> [,8] [,9] [,10] [,11] [,12] [,13] [,14] #> A 1.0000000 0.1751474 0.6013992 0.5136795 1.0000000 1.0000000 1.0000000 #> C 0.3081641 1.0000000 0.3987222 0.5194728 0.6566760 0.8895028 0.6308472 #> G 0.4615689 0.3136375 0.3067372 0.5686857 0.7007176 0.4445597 0.6842762 #> T 0.6408147 0.6368903 1.0000000 1.0000000 0.2172802 0.8877684 0.7344307 #> [,15] [,16] [,17] [,18] [,19] [,20] #> A 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 1.0000000 #> C 0.5410013 0.6158464 0.6555763 0.7516285 0.7736245 0.7317264 #> G 0.7396342 0.6939664 0.6914964 0.7393219 0.7424521 0.7454561 #> T 0.7429453 0.6913035 0.8915726 0.9100964 0.9460106 0.8876650#> Error in as.data.frame.default(motif): cannot coerce class ‘structure("psam", package = "motifStack")’ to a data.frame#> [1] "PSAM.mxr_psam"