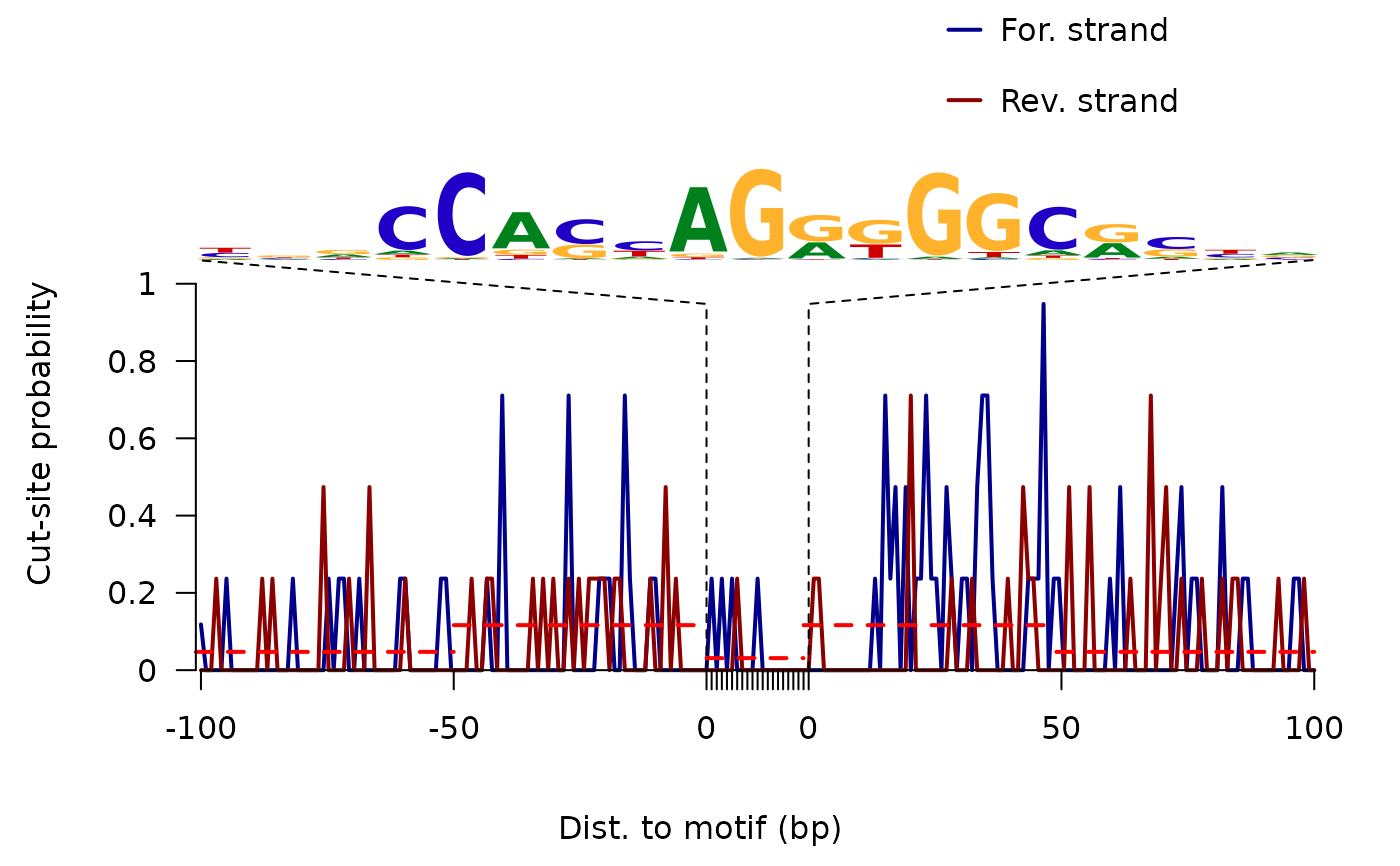
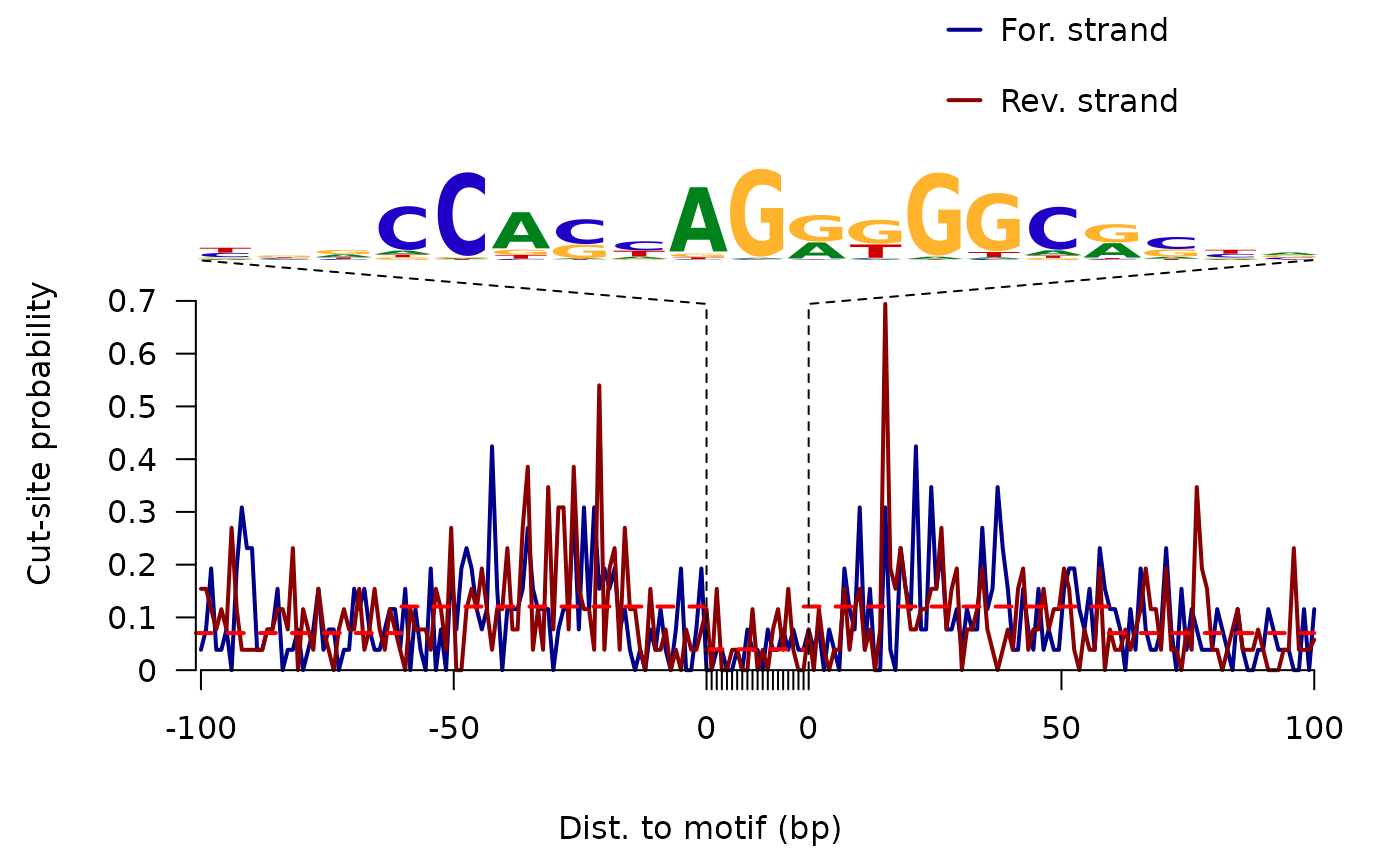plot ATAC-seq footprints infer factor occupancy genome wide
factorFootprints.RdAggregate ATAC-seq footprint for a given motif generated over binding sites within the genome.
factorFootprints( bamfiles, index = bamfiles, pfm, genome, min.score = "95%", bindingSites, seqlev = paste0("chr", c(1:22, "X", "Y")), upstream = 100, downstream = 100, maxSiteNum = 1e+06, anchor = "cut site", group = "strand", ... )
Arguments
| bamfiles | A vector of characters indicates the file names of bams. All the bamfiles will be pulled together. |
|---|---|
| index | The names of the index file of the 'BAM' file being processed; This is given without the '.bai' extension. |
| pfm | A Position frequency Matrix represented as a numeric matrix with row names A, C, G and T. |
| genome | An object of BSgenome. |
| min.score | The minimum score for counting a match. Can be given as a character string containing a percentage (e.g. "95 score or as a single number. See matchPWM. |
| bindingSites | A object of GRanges indicates candidate binding sites (eg. the output of fimo). The GRanges object must have score column in the metadata column. |
| seqlev | A vector of characters indicates the sequence levels. |
| upstream, downstream | numeric(1) or integer(1). Upstream and downstream of the binding region for aggregate ATAC-seq footprint. |
| maxSiteNum | numeric(1). Maximal number of predicted binding sites. if predicted binding sites is more than this number, top maxSiteNum binding sites will be used. |
| anchor | "cut site" or "fragment center". If "fragment center" is used, the input bamfiles must be paired-end. |
| group | Group information for the bamfiles. Default by strand. Otherwise, a factor or vector of characters with same length of bamfiles. The group is limited to 2 groups. |
| ... | xlab, legTitle, xlim or ylim could be used by plotFootprints |
Value
an invisible list of matrixes with the signals for plot. It includes: - signal mean values of coverage for positive strand and negative strand in feature regions - spearman.correlation spearman correlations of cleavage counts in the highest 10-nucleotide-window and binding prediction score. - bindingSites predicted binding sites.
References
Chen, K., Xi, Y., Pan, X., Li, Z., Kaestner, K., Tyler, J., Dent, S., He, X. and Li, W., 2013. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome research, 23(2), pp.341-351.
Author
Jianhong Ou, Julie Zhu
Examples
bamfile <- system.file("extdata", "GL1.bam", package="ATACseqQC") library(MotifDb) CTCF <- query(MotifDb, c("CTCF")) CTCF <- as.list(CTCF) library(BSgenome.Hsapiens.UCSC.hg19) factorFootprints(bamfile, pfm=CTCF[[1]], genome=Hsapiens, min.score="95%", seqlev="chr1", upstream=100, downstream=100)##### Using user defined binding sites ##### bds <- readRDS(system.file("extdata", "jolma2013.motifs.bindingList.95.rds", package="ATACseqQC")) bindingSites <- bds[["Hsapiens-jolma2013-CTCF"]] seqlev <- "chr1" #seqlevels(bindingSites) factorFootprints(bamfile, pfm=CTCF[[1]], bindingSites=bindingSites, seqlev=seqlev, upstream=100, downstream=100)