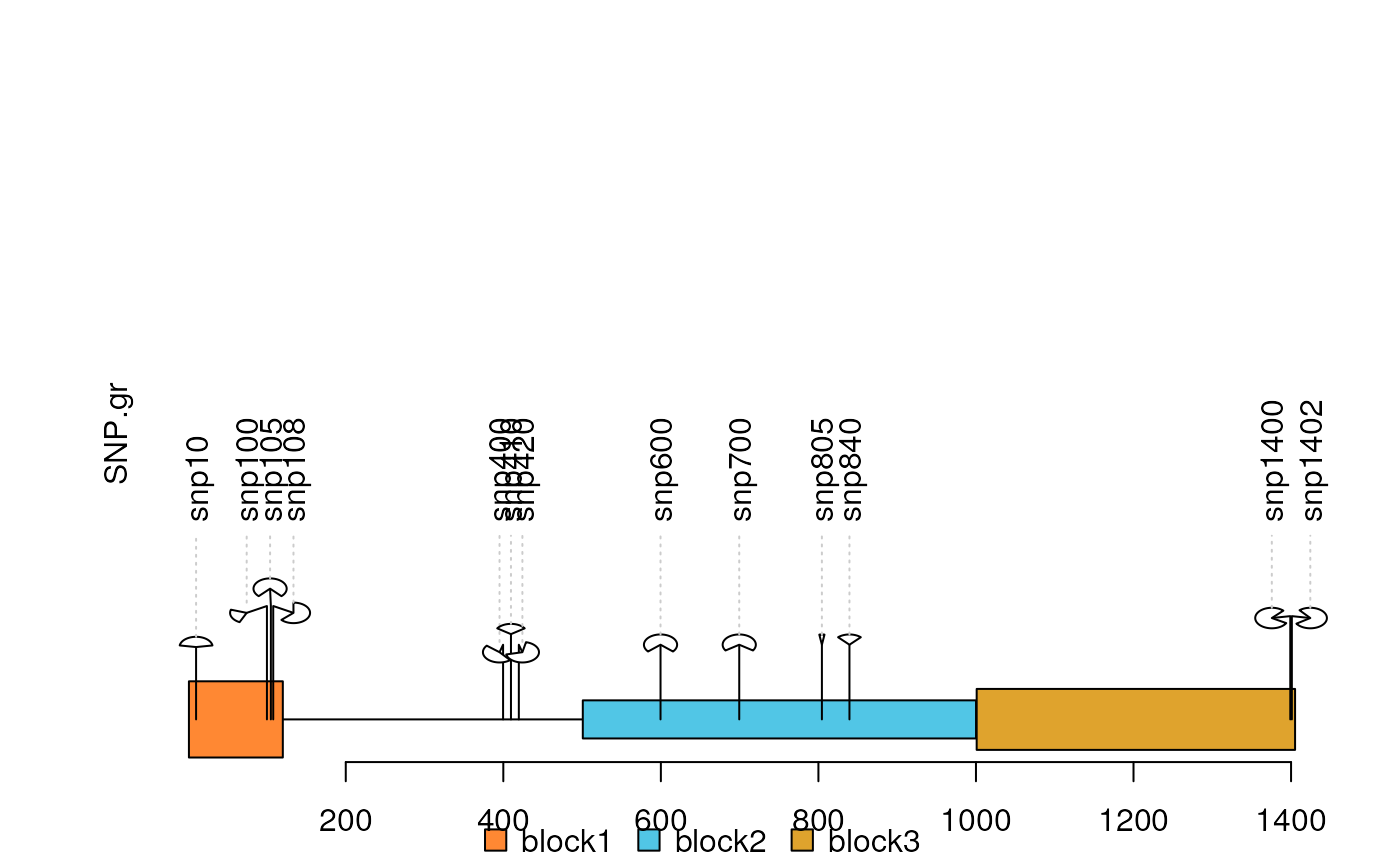
dandelion.plots
dandelion.plot.RdPlot variants and somatic mutations
dandelion.plot(
SNP.gr,
features = NULL,
ranges = NULL,
type = c("fan", "circle", "pie", "pin"),
newpage = TRUE,
ylab = TRUE,
ylab.gp = gpar(col = "black"),
xaxis = TRUE,
xaxis.gp = gpar(col = "black"),
yaxis = FALSE,
yaxis.gp = gpar(col = "black"),
legend = NULL,
cex = 1,
maxgaps = 1/50,
heightMethod = NULL,
label_on_feature = FALSE,
...
)Arguments
- SNP.gr
A object of GRanges or GRangesList. All the width of GRanges must be 1.
- features
A object of GRanges or GRangesList.
- ranges
A object of GRanges or GRangesList.
- type
Character. Could be fan, circle, pie or pin.
- newpage
plot in the new page or not.
- ylab
plot ylab or not. If it is a character vector, the vector will be used as ylab.
- ylab.gp, xaxis.gp, yaxis.gp
An object of class gpar for ylab, xaxis or yaxis.
- xaxis, yaxis
plot xaxis/yaxis or not. If it is a numeric vector with length greater than 1, the vector will be used as the points at which tick-marks are to be drawn. And the names of the vector will be used to as labels to be placed at the tick points if it has names.
- legend
If it is a list with named color vectors, a legend will be added.
- cex
cex will control the size of circle.
- maxgaps
maxgaps between the stem of dandelions. It is calculated by the width of plot region divided by maxgaps. If a GRanges object is set, the dandelions stem will be clustered in each genomic range.
- heightMethod
A function used to determine the height of stem of dandelion. eg. Mean. Default is length.
- label_on_feature
Labels of the feature directly on them. Default FALSE.
- ...
not used.
Details
In SNP.gr and features, metadata of the GRanges object will be used to control the color, fill, border, height, data source of pie if the type is pie.
Examples
SNP <- c(10, 100, 105, 108, 400, 410, 420, 600, 700, 805, 840, 1400, 1402)
SNP.gr <- GRanges("chr1", IRanges(SNP, width=1, names=paste0("snp", SNP)),
score=sample.int(100, length(SNP))/100)
features <- GRanges("chr1", IRanges(c(1, 501, 1001),
width=c(120, 500, 405),
names=paste0("block", 1:3)),
color="black",
fill=c("#FF8833", "#51C6E6", "#DFA32D"),
height=c(0.1, 0.05, 0.08))
dandelion.plot(SNP.gr, features, type="fan")