plot the tracks
viewTracks.RdA function to plot the data for given range
viewTracks(
trackList,
chromosome,
start,
end,
strand,
gr = GRanges(),
ignore.strand = TRUE,
viewerStyle = trackViewerStyle(),
autoOptimizeStyle = FALSE,
newpage = TRUE,
operator = NULL,
smooth = FALSE,
lollipop_style_switch_limit = 10
)Arguments
- trackList
an object of
trackList- chromosome
chromosome
- start
start position
- end
end position
- strand
strand
- gr
an object of
GRanges- ignore.strand
ignore the strand or not when do filter. default TRUE
- viewerStyle
an object of
trackViewerStyle- autoOptimizeStyle
should use
optimizeStyleto optimize style- newpage
should be draw on a new page?
- operator
operator, could be +, -, *, /, ^, %%, and NA. "-" means dat - dat2, and so on. NA means do not apply any operator. Note: if multiple operator is supplied, please make sure the length of operator keep same as the length of trackList.
- smooth
logical(1) or numeric(). Plot smooth curve or not. If it is numeric, eg n, mean of nearby n points will be used for plot. If it is numeric, the second number will be the color. Default coloer is 2 (red).
- lollipop_style_switch_limit
The cutoff value for lollipop style for the 'circle' type. If the max score is greater than this cutoff value, trackViewer will only plot one shape at the highest score. Otherwise trackViewer will draw the shapes like `Tanghulu`.
Value
An object of viewport for addGuideLine
See also
See Also as addGuideLine, addArrowMark
Examples
extdata <- system.file("extdata", package="trackViewer",
mustWork=TRUE)
files <- dir(extdata, "-.wig")
tracks <- lapply(paste(extdata, files, sep="/"),
importScore, format="WIG")
tracks <- lapply(tracks, function(.ele) {strand(.ele@dat) <- "-"; .ele})
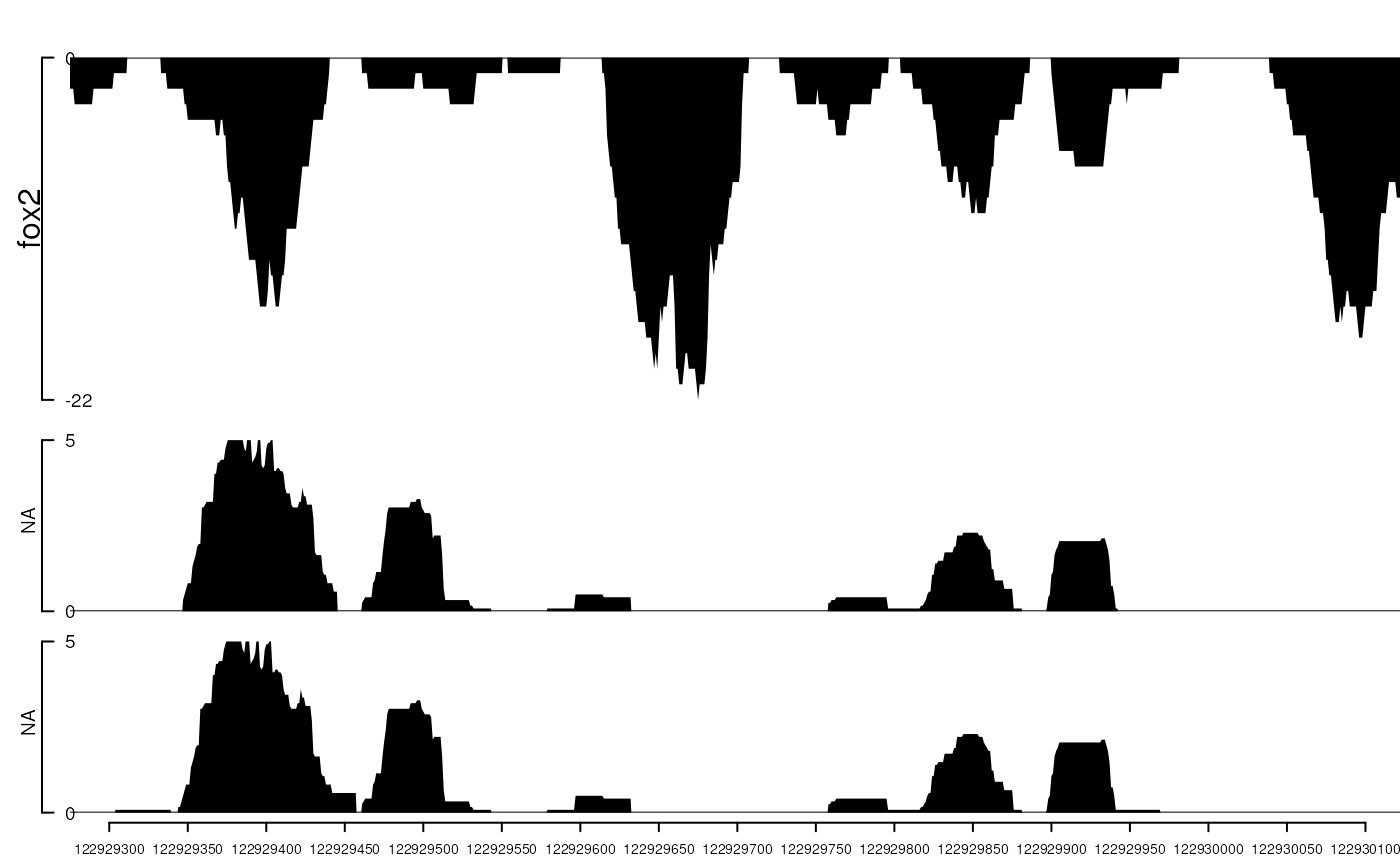
fox2 <- importScore(paste(extdata, "fox2.bed", sep="/"), format="BED")
dat <- coverageGR(fox2@dat)
fox2@dat <- dat[strand(dat)=="+"]
fox2@dat2 <- dat[strand(dat)=="-"]
gr <- GRanges("chr11", IRanges(122929275, 122930122), strand="-")
viewTracks(trackList(track=tracks, fox2=fox2), gr=gr, autoOptimizeStyle=TRUE)